Nuclear Magnetic Relaxation Dispersion of Water-Protein Systems
Nuclear Magnetic Relaxation Dispersion (NMRD) has been carried out on the investigation of molecular motions on water-protein systems. The spin-lattice relaxation rate constant (R1) has a certain proportional relationship to the spectral density function, which is a function of the nuclear Larmor frequency. Therefore, utilizing the field-cycling NMR technique can obtain such an NMRD profile on different studied systems.
This issue we pick a few examples for a journal tour of the recent NMRD studies on water-protein systems. NMRD has been applied to probing different time scales of dynamics. Here, we choose an article using NMRD to determine microsecond reorientation correlation times in protein solutions. The other article is to probe pico-second dynamics by utilizing paramagnetic contribution in water-protein systems.
Reorientation correlation times in protein solutions are key determinants for feasibility and quality of NMR experiments. Yet, their accurate estimate is not easy, especially in the case of very large proteins. We show that nuclear magnetic relaxation dispersion (NMRD) can accurately determine reorientation times up to the microsecond range. A theoretical description for the analysis of the NMRD profiles is provided, and the protein reorientation time is shown to be provided by the longest correlation time among those needed to reproduce the experimental profile. Measurements are performed using samples of the archaeal proteasome double ring α7α7 and of αB-Crystallin in glycerol solutions.
Water-Protein Systems
Experimental Determination of Microsecond Reorientation Correlation Times in Protein Solutions
Ravera et al, J. Phys. Chem. B. 2013, 117, 3548−3553
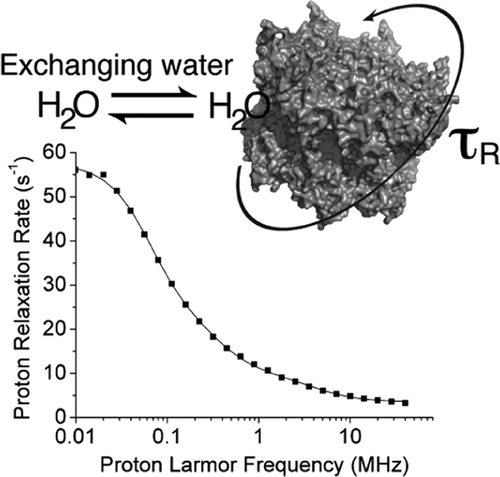
Reorientation correlation times in protein solutions are key determinants for feasibility and quality of NMR experiments. Yet, their accurate estimate is not easy, especially in the case of very large proteins. We show that nuclear magnetic relaxation dispersion (NMRD) can accurately determine reorientation times up to the microsecond range. A theoretical description for the analysis of the NMRD profiles is provided, and the protein reorientation time is shown to be provided by the longest correlation time among those needed to reproduce the experimental profile. Measurements are performed using samples of the archaeal proteasome double ring α7α7 and of αB-Crystallin in glycerol solutions.
Water-proton-spin–lattice-relaxation dispersion of paramagnetic protein solutions
Diakova et al, J. Mag. Res. 2011, 208, 195-203
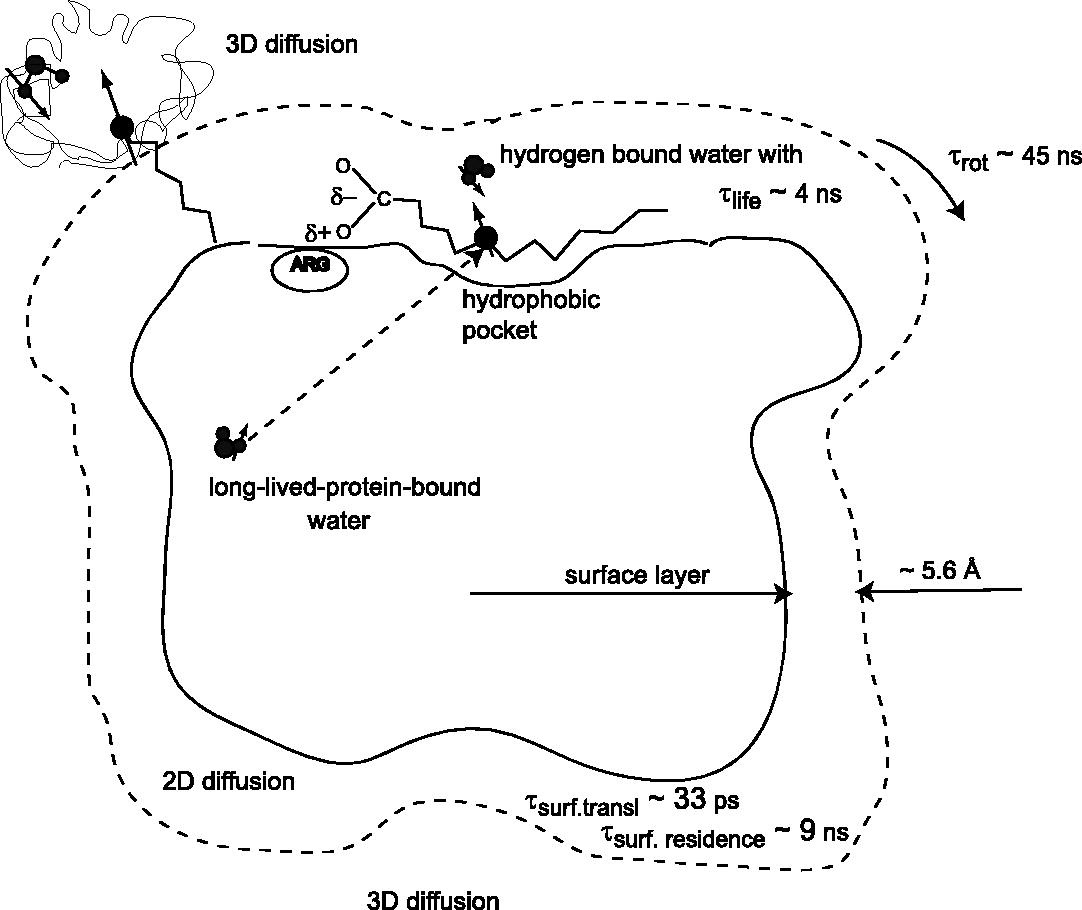
The excess water-proton-relaxation efficiency is examined for paramagnetic proteins. The model for the effects of 2-dimensional diffusion is developed but is insufficient to account entirely for the excess. The effects of long-lived-protein -bound water is shown to be important when the paramagnetic center has a long T1e.
Analysis Method
Model-Free Analysis of Stretched Relaxation Dispersions
Halle et al, J. Mag. Res. 1998; 135(1): 1-13.
Nuclear magnetic relaxation dispersion (NMRD) measurements can provide valuable information about the dynamics and structure of macromolecular solutions and other complex fluids. A large number of1H NMRD studies of water in concentrated protein solutions and in semisolid biological samples have been reported. The observed dispersion usually extends over a wide frequency range and then cannot be described by a Lorentzian spectral density function. We propose here a model-free approach for analyzing such stretched dispersion profiles. Unlike the traditional empirical fitting procedures, the model-free approach is based on rigorous theory and produces parameters with well-defined physical significance. The model-free approach is validated with the aid of synthetic relaxation data, showing that it is robust and accurate, and is then applied to new water1H NMRD data from solutions of the protein bovine pancreatic trypsin inhibitor (BPTI). By separating the static and dynamic information content of the relaxation dispersion, the model-free analysis shows that the dramatic salt effect observed in BPTI solutions is due almost entirely to a slowing down of protein rotation with little change of protein structure. An analysis of the same data in terms of the empirical dispersion function used in most1H NMRD studies leads to a qualitatively different picture. We demonstrate that this widely used dispersion function is unphysical and that its parameters do not have the physical meaning usually ascribed to them.